By ART LATHAM
Despite the 20-knot northeast wind whipping up the Spanish moss and gusting across the sandy pine-and-scrub flats outside the bunkhouse window at the isolated marine research lab, the lunchers at the communal table inside are in fine fettle.
Much of the mealtime banter at the lab near Aurora is science-related, but it’s easy to tell that the two men tormenting each other — Craig Sullivan and Ron Hodson — actually are good friends.
The two longtime North Carolina Sea Grant researchers obviously enjoy hurling salty epithets across their plates or muttering asides into their sodas. Their good moods are infectious, and they have reason to celebrate. After more than 20 years of studies that another team member tagged “extreme research,” the two scientists are nearing a lifelong research goal.
Sullivan, a North Carolina State University biologist and his team, including Hodson who continues to participate in research since retiring as North Carolina Sea Grant director in 2006, recently completed a genomic map of the striped bass. This is the first map for any non-salmonid marine species farmed in the United States, and, as such, is a big deal.
“Maps” are crucial to the success of selective breeding, in this case, the commercial breeding of the hybrid striped bass. Crossing, or interbreeding, two wild bass types — striped and white — produces the hybrid striped bass, a major component of North Carolina’s multimillion-dollar aquaculture industry.
Scientists use both linkage maps and sequencing to produce portraits of a genome.

These huge tanks contain domesticated bass breeding stock. Photo by Becky Kirkland.
A map, which identifies the locations of a series of landmarks in a genome, is less detailed than a sequence, which spells out the order of every DNA base in the genome. Scientists use the landmarks, or markers, to properly assemble a genome sequence. The markers also serve as clues about where to find regulatory sites that turn genes on and off, and the genes themselves.
“What we’ve done here is to create signposts that we can recognize,” Sullivan says. “You can’t get there without a map. The next step is to acquire the complete genetic code for striped bass, a step analogous to the Human Genome Project, which has given us the ability to look at and predict various illnesses and to identify a whole host of new targets for drug therapy and much more.”
The team started out with nothing. “It’s analogous to going into the jungle to catch guinea fowl to create a poultry industry. But we pioneered the commercial production of hybrid striped bass in North Carolina farm ponds in the late 1980s. The industry has since spread nationwide,” he continues.
“Twenty years later, we’ve established a national breeding project here at the North Carolina State University Pamlico Aquaculture Field Laboratory,” he says. “This lab is the center of the striped bass universe, and our diverse stock came from every native stock in North America: Canada, the Hudson River, Chesapeake Bay, the Roanoke River, the Santee-Cooper reservoir and the Gulf coast, among other sources.
In the past two decades, the researchers have developed reliable techniques for hatchery propagation of the fish and selectively bred and domesticated four generations of striped bass and eight of white bass. The effort involves partnership with Adam Fuller and his associates at the U.S. Department of Agriculture (USDA) Stuttgart National Aquaculture Research Center in Arkansas, where duplicate stocks of the fish are being bred.
“Now we’re starting to distribute to producers,” says Sullivan, whose most recent Sea Grant funding includes a mini-grant to study the genetic determinants for striped bass egg quality.
And speaking of producers, also at the lunch table is Lee Brothers, who provided a three-acre pond at his Carolina Fisheries near Aurora to host the first pond-reared hybrid striped bass demonstration.
“You can’t imagine the dollars and the work this research has saved me,” says Brothers, a third-generation Beaufort County farmer who was growing soybeans, corn and tobacco when economic hard times hit. He is one of several commercial seafood producers in North Carolina.
“I had to do something on the farm,” he says. “I had ponds built and was going to do hybrids when I found out about this program. They’ve trained a lot of people in breeding striped bass, which are hard to breed. I got interested in what they were doing, helped collect a couple of ‘stripers,’ then got interested in watching them spawn.
“We got our first fingerlings in 1987 and sold our first crop in the winter of 1988-89. The first harvest was 70,000 pounds. Now it’s about 500,000 pounds,” he says.
Brothers, who raises “a few” fingerlings — younger fish — for other producers, says each year’s crop is improving, but adds, “Our mainstay is food fish. That’s where the money is.”
He’s now rearing his own white bass broodstock, and is ready to start with striped bass broodstock, the sooner the better, he says. And well he should be.
Due in large part to work funded by North Carolina Sea Grant and partners, North Carolina’s striped bass industry burgeoned into high-value status in the past decade. Sold live to ethnic Asian markets and frozen to upscale restaurant markets, hybrid striped bass sales already are rebounding from the recession, Sullivan notes.
In fact, NC State statistics show that in 2009, 12 North Carolina hybrid striped bass producers reported a 2.4-million-pound harvest from 763 pond-acres. The reported farm-gate value — a farmer’s net income from sale of a product — was more than $9 million. Also, four hatcheries produced almost five million fingerlings, for a farm-gate value of $1.2 million.
“At some point,” Brothers says, “I’m going to have to build a new building and tanks to hold these stripers, because it’s a short spawning season and they can’t take the summer heat. I’m ready to take the fish whenever they’re ready to give them to me.”
EXTREME RESEARCH
Lunch break over, the researchers brave the wind to gather around a giant, uncovered tank to seine out a few female striped bass. They’re checking to see if the females are showing a high enough egg count to spawn successfully. Andy McGinty, site supervisor and Mike Hopper, research specialist, wrestle with a seine net to capture two good-sized, thrashing fish.
While Brothers noted some of the challenges involved in developing hybrid striped bass, exactly how “extreme” can scientific research be? It takes but a few minutes outdoors to answer that question.
Try leaning over a huge fish tank in winter, your hands chapping to rough flakes, your nose and ears frozen into popsicles as the frosty wind whips across the Pamlico River to the north and right up your shirtsleeves.
Or sweating heavily while clad in a waterproof outfit that makes you resemble an iconic fisherman, while you jockey large, reluctant, splashing striped bass from the tank in a wind with a chill factor that could stun an Aleut. Or how about pulling back-to-back, 24-hour shifts during the springtime spawning season?
You do all this so you can then perform such delicate operations as sticking a tube into the oviduct of a female striped bass, withdrawing and measuring the growth of eggs she’s producing: 750 microns diameter on this winter day. They’ll be over 1,000 microns when she’s ready to breed, Sullivan says.
What’s the payoff for 20 years of probing the ovaries and testes of splashy, slippery stripers?
For one thing, the first linkage map for any of the Morone species, temperate-water basses that swim off North Carolina’s ocean shores and in its rivers, brackish bays and inland waters. So far in the United States, this mapping has only been accomplished for a handful of farmed fishes: Atlantic salmon, rainbow trout, channel catfish, tilapia and now, striped bass.
“A genetic map is crucial to selective breeding,” says Sullivan. “It’s the backbone, the structure on which we will hang the genome sequence of the striped bass.”
Variants of the markers on such maps also help scientists identify future broodstock that will produce offspring with better growth, feed conversion and yields.
NITTY-GRITTY LAB TEAMWORK
Working with collaborators including Caird Rexroad, Sullivan’s team developed about 500 new microsatellite DMA markers (MSMs) as the first step toward mapping the striped bass genome. Researchers use MSMs to identify offspring produced by each parental pair involved in a series of crosses to provide information on the genetic basis of specific traits.

Clockwise, from back left: Craig Sullivan, Ron Hodson, Andy McGinty and Mike Hopper conduct research at North Carolina State University’s Pamlico Aquaculture Field Laboratory. Photo by Becky Kirkland.
Rexroad is at the USDA National Center for Cool and Cold Water Aquaculture in Kearneysville, W. Va. Other partners in the mapping project, which was funded by the National Oceanic and Atmospheric Administration’s Marine Aquaculture Initiative, include Charlene Couch of NC State’s Genetics Department and a former North Carolina Sea Grant fellow; Sixin Liu, molecular biologist at the USDA center; and Kimberly Reece and Jan Cordes, geneticists at the Virginia Institute of Marine Science.
Microsatellite DNA marker development also depended on the DNA sequencers at NC State’s College of Agriculture and Life Sciences Genomic Sciences Laboratory, Sullivan says.
The NC State researchers created two striped bass “mapping families,” genotyped the fish at several marker positions and assessed their physical characteristics to ensure they were genetically and physically variable enough to map. The Virginia scientists genotyped the parents and offspring at 293 more marker positions, identifying which of several marker variants each fish had at each position. The USDA scientists then analyzed the genotype data to construct the final linkage map, on which markers are positioned relative to one another based on how well their variants remain together or are “linked” during production of gametes and offspring.
The next research step will be to evaluate whether variants at some marker positions are associated with high performance of the fish with respect to important quantitative traits, such as growth or body shape. Such markers exist at points called quantitative trait loci (QTL). Mapping helps researchers use QTLs to move from selection by phenotype — an individual’s measurable physical characteristics, often evident by outward appearance — to selection by genotype, which is an organism’s genetic makeup, in this case the marker variants present at certain positions on the map.
“Obviously we’re mapping genomes for a reason,” Sullivan notes.
Several reasons, actually, such as:
• Helping researchers discover what genes affect biochemical processes underlying important fish traits, such as growth or body conformation.
• Researching whether the genes are altered when wild fish are domesticated in captivity.
• Genetically “branding” the special NCSU-WB1 strain of white bass broodstock for distribution, urgently needed now because Lake Erie, once a major source of wild female white bass for hybrid striped bass production, closed a few years ago to collectors.
That loss meant increasing pressures on the hybrid striped bass industry to develop or adopt reliable domesticated broodstock. Somebody had to address that situation. Sullivan and his team picked up the challenge, and they got the job done.
Hodson — who arrived at North Carolina Sea Grant in 1981, was named interim director in 1997 and full director in 1998 — recalls various steps. “I came on the scene when Sea Grant was just starting to back the hybrid striped bass program. By 1987, we were ready for commercial-scale demonstrations.”
That initial research focused on transferring technology for in-pond hybrid striped bass production on farms, so researchers stocked ponds with lingerlings in exchange for a year’s worth of data from growers.
“We have worked to get something going with selective breeding,” Hodson says. “You’ve got to get the broodstock first.”
Fast-forward to 2011 and the research continues, as Sullivan compares offspring from crossbreedings, thanks to a new N.C. Fishery Resource Grant project.
SHOT ACROSS THE BOW
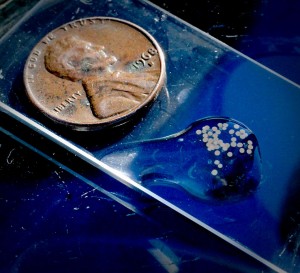
Tiny striped bass eggs are dwarfed by a penny. Photo by Becky Kirkland.
The research has provided many rewards.
In December, Sullivan was named a fellow of the American Association for the Advancement of Science. The association recognized his work in “vertebrate reproductive biology, advancing knowledge of oogenesis in fishes and establishing striped bass farming as a major form of aquaculture.”
Sullivan — who with his team presented their most recent genome mapping in San Diego, Calif., at the international Plant and Genome Conference in January — appreciates the epic nature of genomic research.
“It’s the odyssey that I think is interesting,” he says, “that we’ve come to this from capturing wild animals to selective breeding of the fish and mapping the genome. It’s been a wild ride.
“But we’ve fired the first shot across the bow, taken the first step in what will be the Striped Bass Genome Project.”
HSB PROJECT FUNDING SOURCES
The broodstock development effort for both striped and white bass has been supported by North Carolina Sea Grant and other programs, including:
• North Carolina State University, North Carolina Agricultural Research Service
• University of North Carolina, Office of the President, Genomic Sciences Grant Program
• U.S. Department of Agriculture’s National Institute of Food and Agriculture (formerly USDA-CSREES)
• U.S. Department of Commerce, National Oceanic and Atmospheric Administration (NOAA) Marine Aquaculture Program
WANT TO LEARN MORE? TRY THESE WEBSITES:
Craig Sullivan research
https://marinesci.ncsu.edu/people/sullivan/
Genome News Network
http://www.genomenewsnetwork.org/
Hybrid striped bass in N.C. updates
www.ncaquaculture.org/documents/2009brochure.pdf
NOAA Aquaculture Program
https://www.fisheries.noaa.gov/topic/aquaculture
North Carolina State University graduate program in genomics
U.S. Department of Agriculture’s Agricultural Research Service: genome-related term definitions http://www.ars.usda.gov/is/AR/archive/aug08/genotypic0808.htm
This article was published in the Winter 2011 issue of Coastwatch.
For contact information and reprint requests, visit ncseagrant.ncsu.edu/coastwatch/contact/.
